1.1 SNP tables
DoGSD database summarizes the SNP number by chromosome of 127 samples (11 wolves and 116 dogs) and dbSNP(version 146), both with individual and nonredundant resolutions. Click a number, and you will see the whole SNP list of a given chromosome.
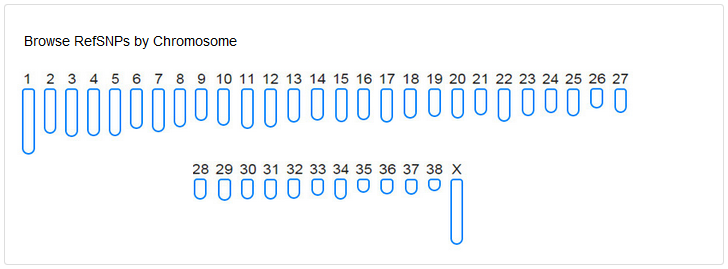
1)Reference SNP list
The #Reference SNP will redirect users to a reference SNP table that shows the general SNP information of all samples. If one SNP locates in a gene, the gene id, consequence type and SNP effect will be displayed. Clicking the RSNP ID will redirect to the detail information page of this position, and clicking the JBrowse link will open the JBrowse view panel.
On this page, users can use advance operations including “Filter”, “Edit Columns” and “Choose Individual”.

2) The reference SNP detail information
The reference SNP is non-redundant SNP collection by combining all the individuals in the whole genome scale, the detailed information is used to display the whole information of a given SNP site, which mainly contains general information, gene information, individual genotype and population analysis.
General Information will list the gene ID, version number, chromosome and position, and SNP class of a given SNP. A link in RefSNP of version 1 will also be given.
In gene information, the gene model table shows the gene list where SNP are located. The transcript and protein table will show the transcript and protein related to the reference SNP. Clicking on a Gene ID will redirect to the detail information of that gene in DoGSD, and clicking on the transcript or protein accession will redirect to the Ensembl database.
In individual genotype, the SNP of all samples at this position will be listed.
In population analysis, the genotype frequencies and allele frequencies will be displayed.

3) Individual SNP list
Clicking on a SNP ID will redirect to the detail information page of this SNP.

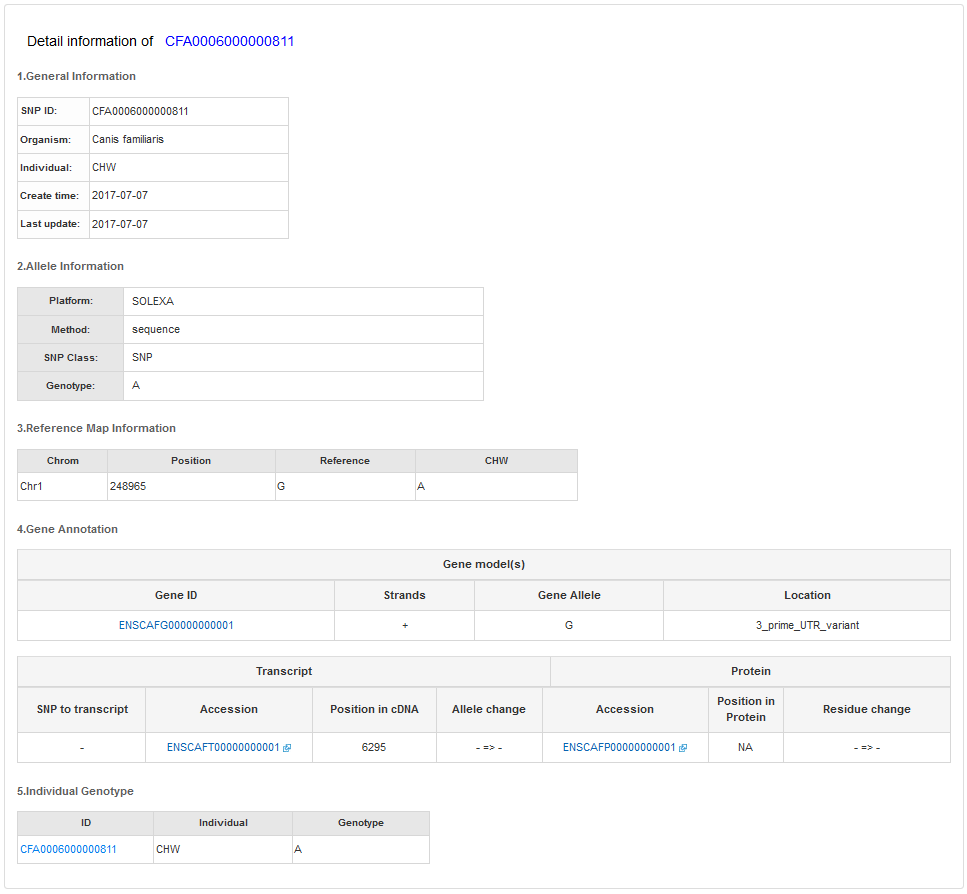
4) SNP detail information
The SNP detail information is for users to view the SNP information of a specific individual at a given position. The information are presented in five categories.
a) The General Information will show SNP ID, individual, SNP loaded time and the NCBI rs id of a SNP.
b) The Allele Information will show the individuals genotype information.
c) The Reference Map Information will show the position, derived and reference alleles.
d) The Gene Information will show the information of related gene, transcript and proteins of a SNP.
e) The SNPs of other individuals in this position will show the genotype information of other individuals.
5) Gene detail information
The gene detail information shows SNPs located in the corresponding gene.

1.2 SNPs in Gene
SNP in Gene Statistics will list SNPs located in the gene and coding regions. According to the protein sequence, we split the SNP list into synonymous and non-synonymous. Clicking the number in the columns will show the whole SNP list in the corresponding chromosome.
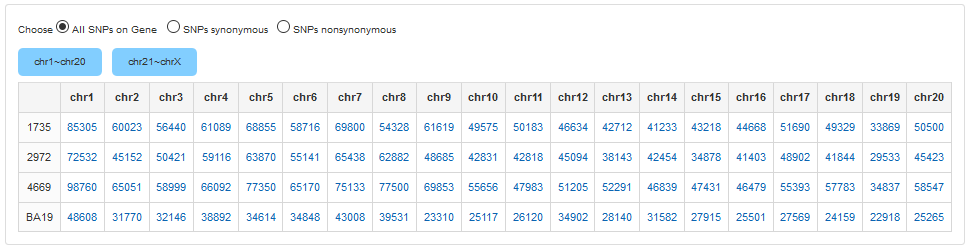
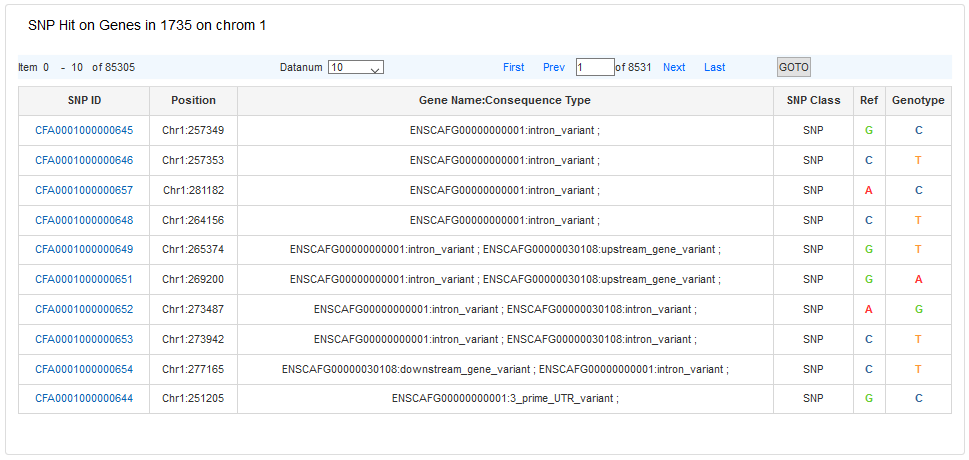
The # All SNPs on Gene list is used to view SNP information located in different gene region. Clicking a SNP ID will redirect to the detail information page of this SNP.
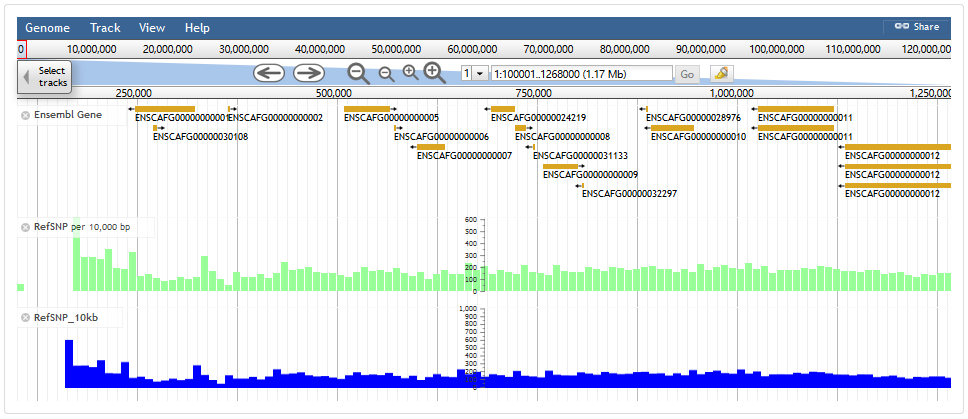
1.3 Browse on Chromosome
DoGSD database uses JBrowse to visualize several data types, including the SNP, gene, transcript and the density information of SNP/300kb, allele frequency. Users would need to choose interested tracks in the Select Tracks menu, and a region of interest.
Users can search SNP in DoGSD, including searching SNPs in single individual or comparison of SNPs in two or more individuals. Users can set several filters, including location in chromosome, SNP class (mainly SNP), SNP location in gene region and genotype. We describe the search result of single individual and compared individuals separatly.
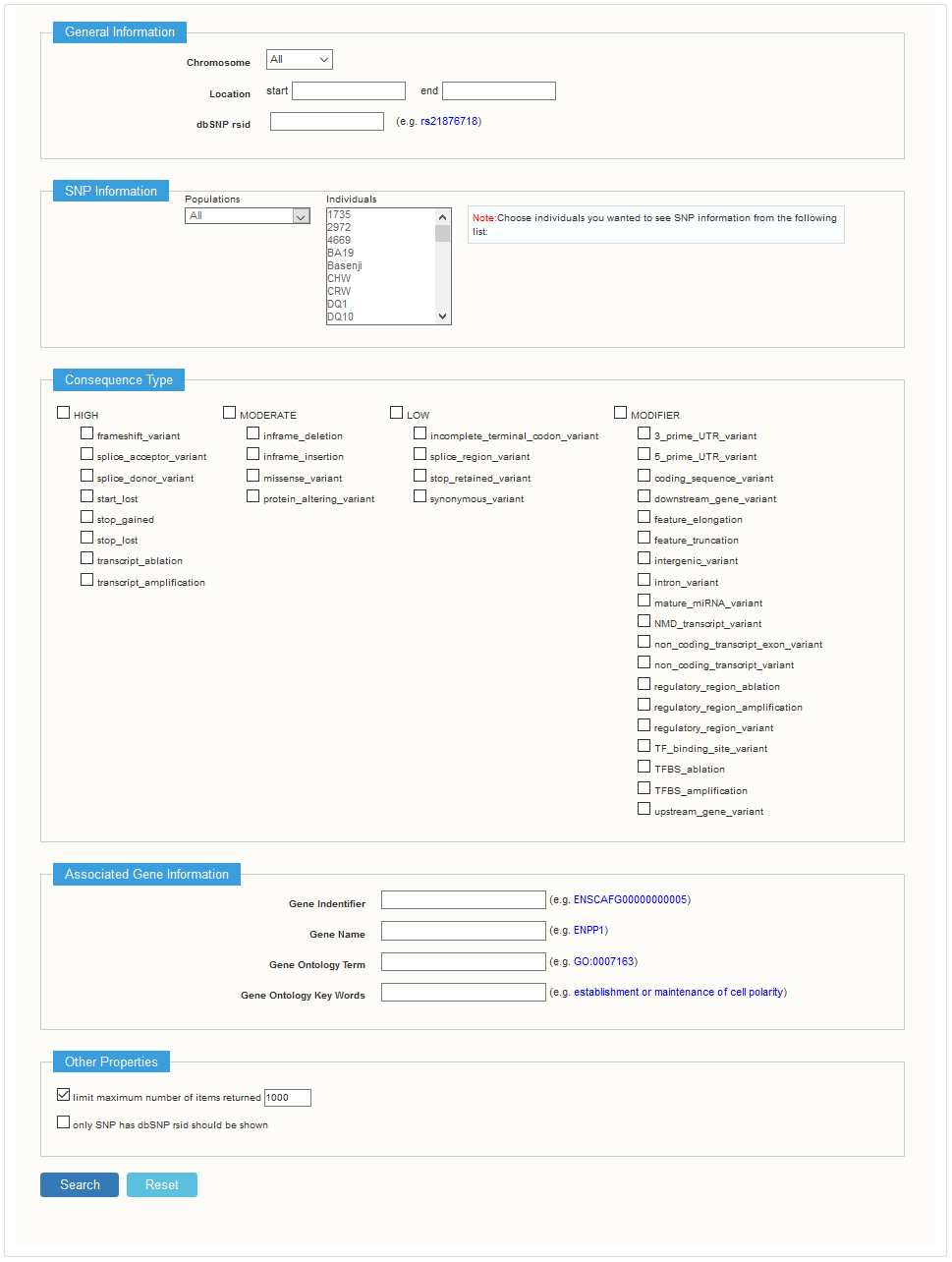
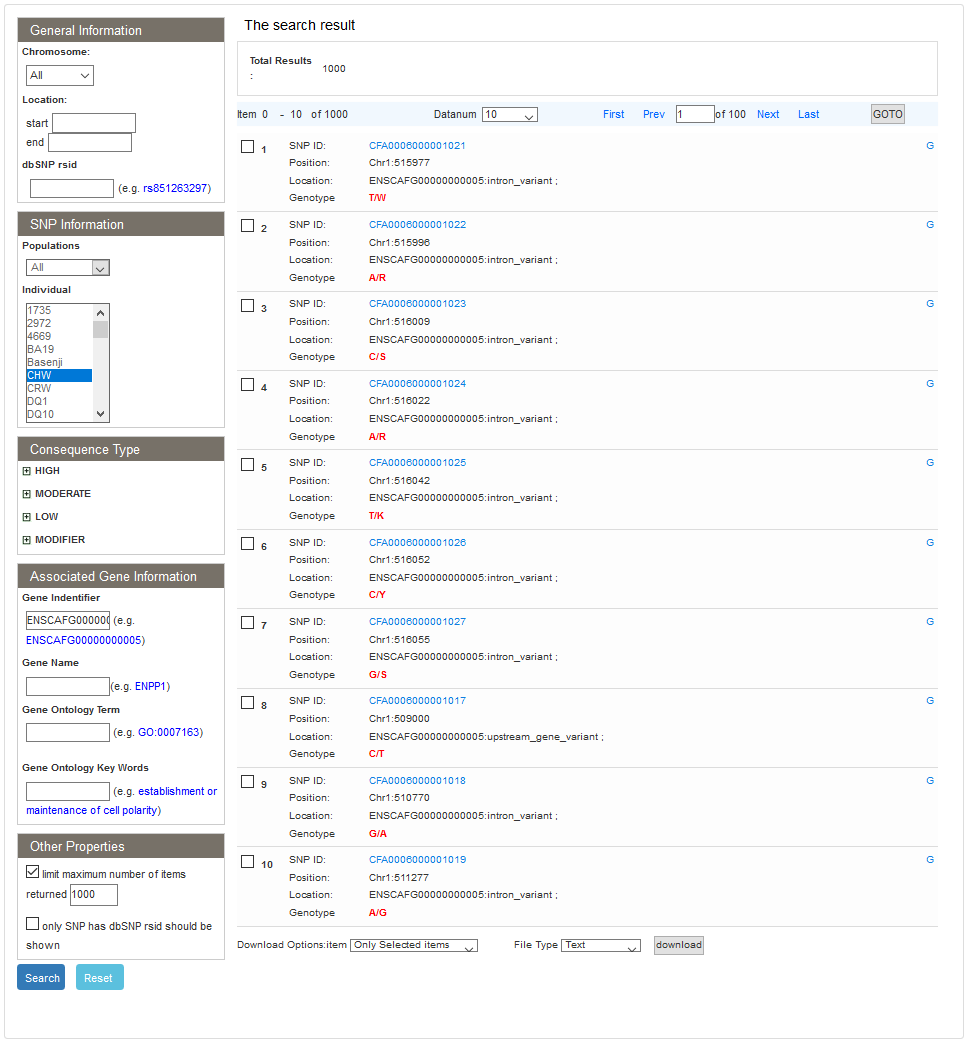
2.1 Single individual search
Users can view SNP data of each individual by choosing the "Show SNP information in individuals" in the search page. On the single search result page, users can view the search conditions, and the total number of SNP results. The SNP list results are listed below, which includes SNP ID, position, location (if SNP is located in gene, the gene name will be show) and flank sequence (upstream and downstream of 30bp at this position). Users can click the GBrowse link to view the SNP in the GBrowse panel. And for detailed information, users can click the SNP ID link.
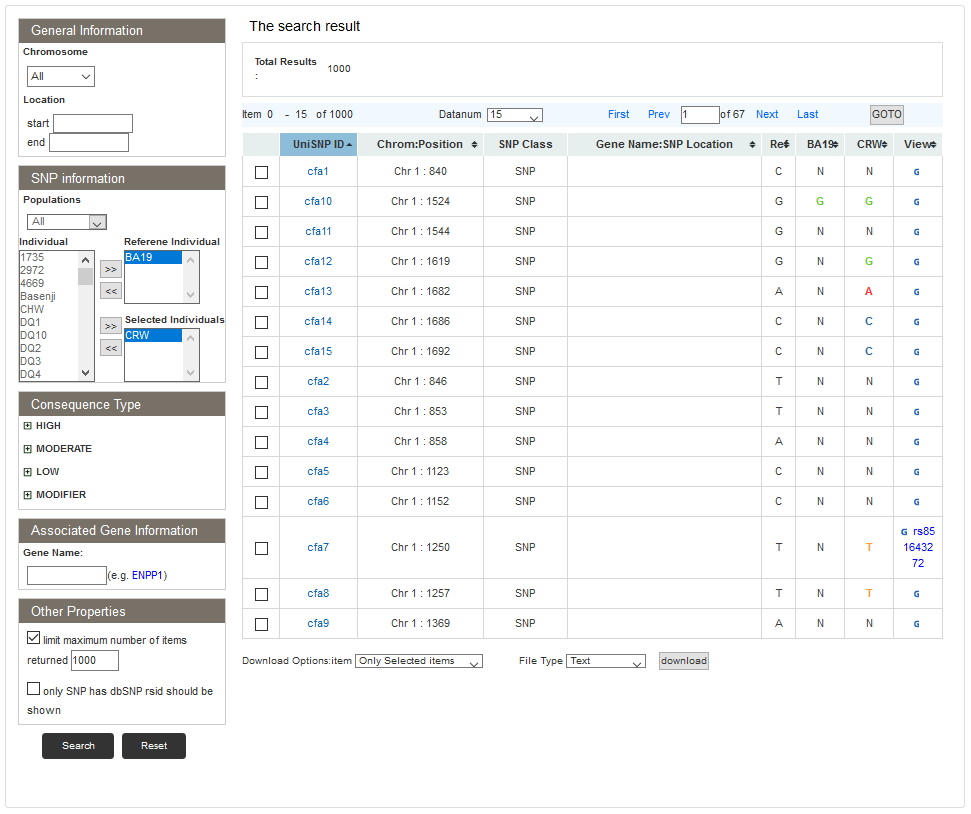
2.2 Compared individuals search
Users can view SNP information of two or more individuals by choosing "Show SNP information by individuals comparisons" in the search page. On the compared individuals search result page, the SNP list will show SNPs in selected individuals based on SNP location of the reference individual. The UniSNP ID refers to the reference SNP, and the columns of the individuals following the “Ref” genotype column corresponds to the order of selected individuals.
Users can download the SNP, bam and fastq files of specific samples from ftp://download.big.ac.cn/pub/dogsd/

