Introduction
1.1 What is LncRNAWiki 2.0?
LncRNAWiki 2.0 is devoted to community curation of human long non-coding RNAs (lncRNAs) to provide a comprehensive and up-to-date resource of functionally annotated lncRNAs.
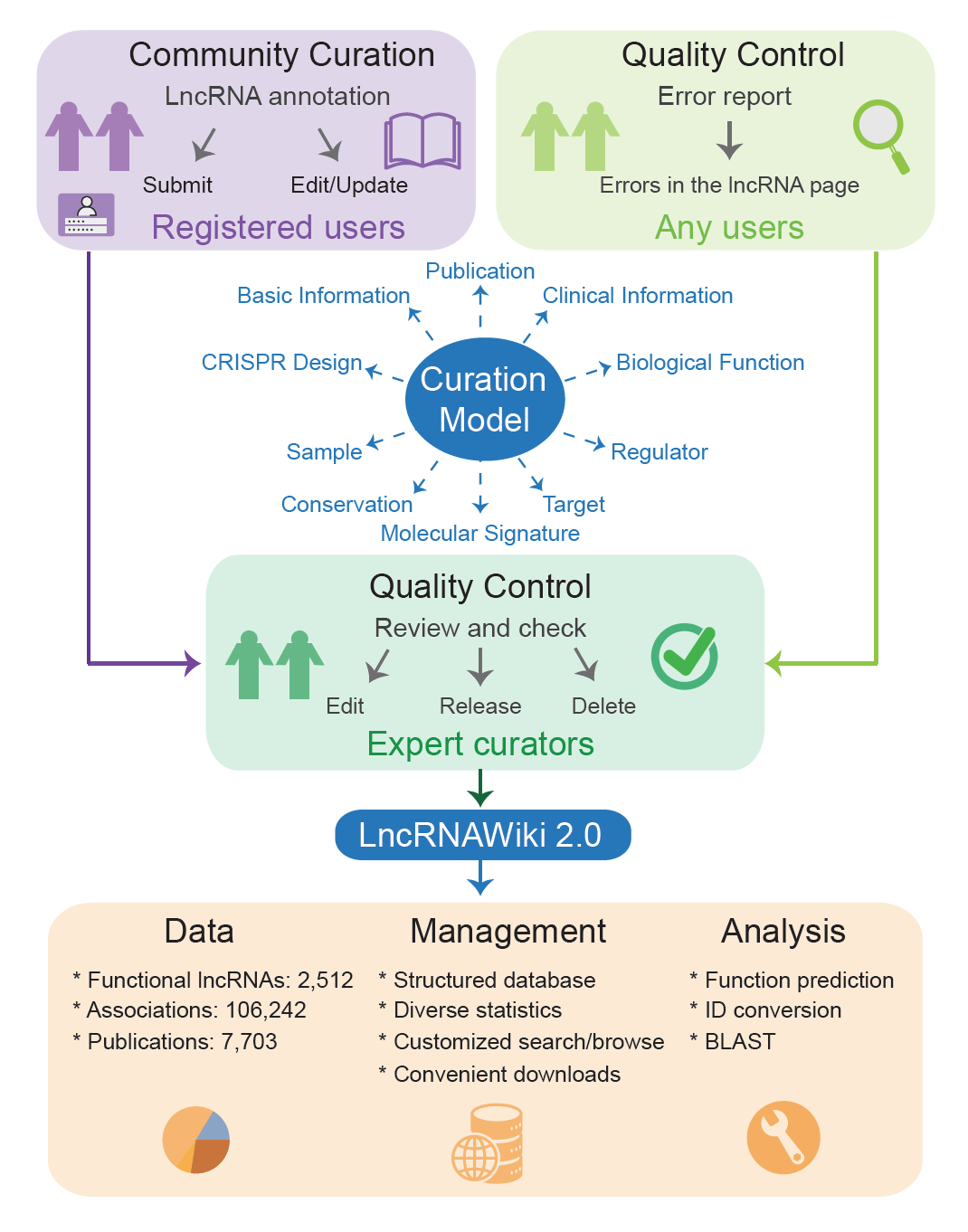
It adopts a standardized curation model that incorporates a wide range of annotations, invloving 10 sections, namely, basic information, publication, conservation, experimental sample, clinical information, biological function, molecular signature, regulator, target, and CRISPR design. Curators (registered users) submit/edit annotations based on the curation model. Also, curation quality is well controlled by recruiting experts to review the curation results and allowing any user to report errors.
Based on the informative curation model, LncRNAWiki 2.0 incorporates a large number of human lncRNAs and their associations with experimental evidence. To enable community curation and provide associated functionalities, LncRNAWiki 2.0 is implemented based on MySQL/Java and significantly improved in database system to facilitate data management and analysis. Friendly web interfaces are accordingly developed to ease data management, including data submission, edit, review, error report, as well as browse, search, download and statistics. Moreover, it provides several online tools for lncRNA ID conversion, sequence search and function prediction.
1.2 What is the difference between LncRNAWiki 1.0 and 2.0?
LncRNAWiki, a wiki-based database for community curation of human lncRNAs, was originally published in the NAR 2015 Database Issue. It has been rapidly expanded by incorporating more experimentally validated lncRNAs. Since it was built based on MediaWiki as its database system, it fails to manage data in a structured way and is ineffective to support systematic exploration of lncRNAs. Here we present LncRNAWiki 2.0, which is significantly improved with enhanced curation model and database system.
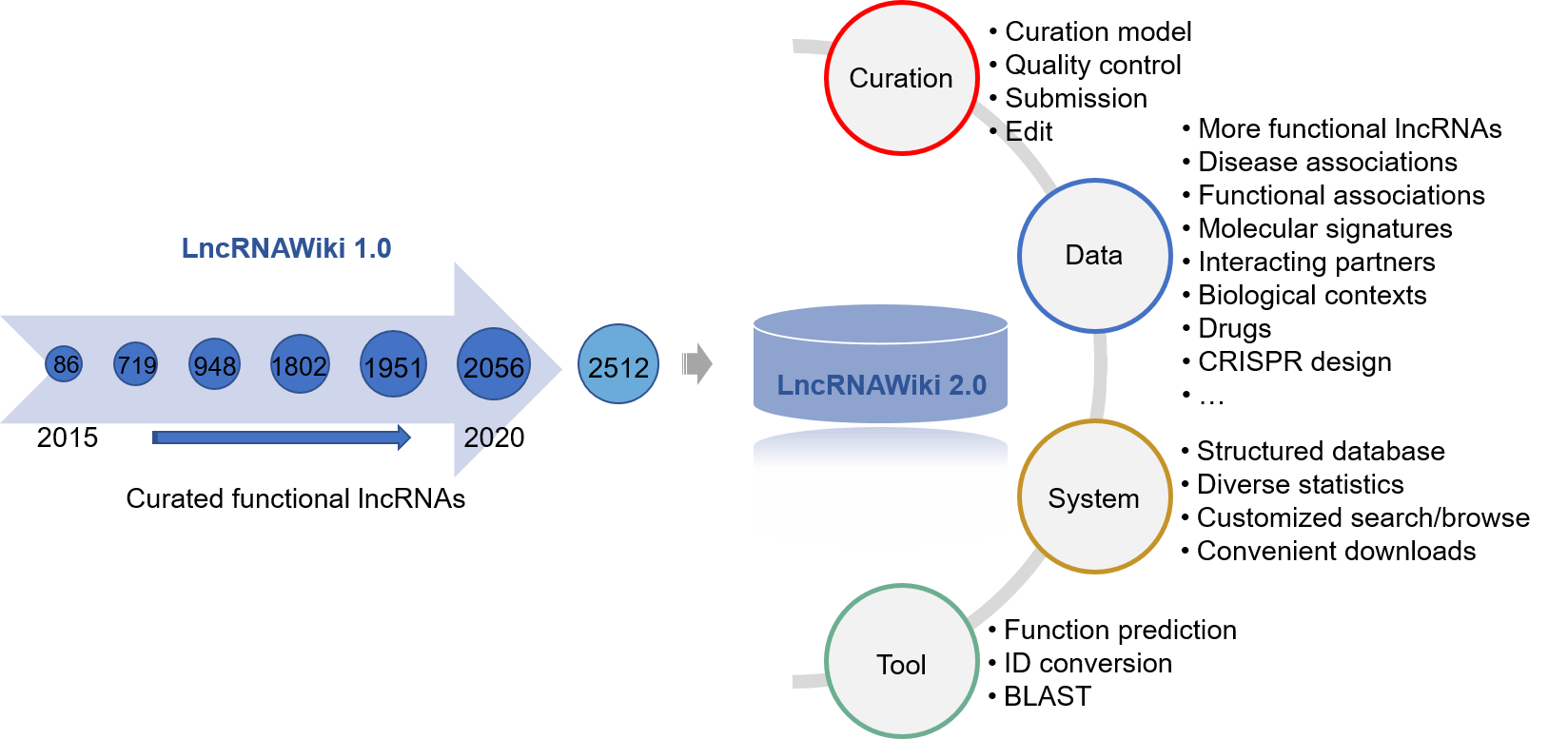
Compared with the first version, LncRNAWiki 2.0 has significant changes and improvements as follows.
- LncRNAWiki 2.0 provides a standardized curation model that includes more informative and essential annotation items for lncRNAs and their associations.
- Curation quality is well controlled by recruiting experts to review the curation results and allowing any user to report errors.
- LncRNAWiki 2.0 is built based on MySQL/Java and thus is capable to organize all contents in a structured manner.
- LncRNAWiki 2.0 provides more user-friendly web interfaces to facilitate data data curation, retrieval and visualization.
- LncRNAWiki 2.0 is equipped with popular online tools to help users identify lncRNAs with potentially important functions.
1.3 What is the difference between LncRNAWiki 2.0 and other knowledgebases?
Different from the existing lncRNA knowledgebases (e.g., LncRNADisease, Lnc2Cancer, LncRNA2Target, LncSLdb, D-lnc) that rely on expert curations with specific curation models in disease association, interaction, subcellular localization, development and etc., LncRNAWiki 2.0 is devoted to efficient community curation, comprehensive and up-to-date annotations, as well as stringent quality control. It features community curation functionality with standardized curation model and friendly web interfaces for information submission, edit, review and error report.
1.4 What kind of the curation model does LncRNAWiki 2.0 provide?
LncRNAWiki 2.0 is devoted to community curation of human lncRNAs based on a structured curation model, which consists of 10 sections covering basic information, publication, conservation, experimental sample, clinical information, biological function, molecular signature, regulator, target and CRISPR design.
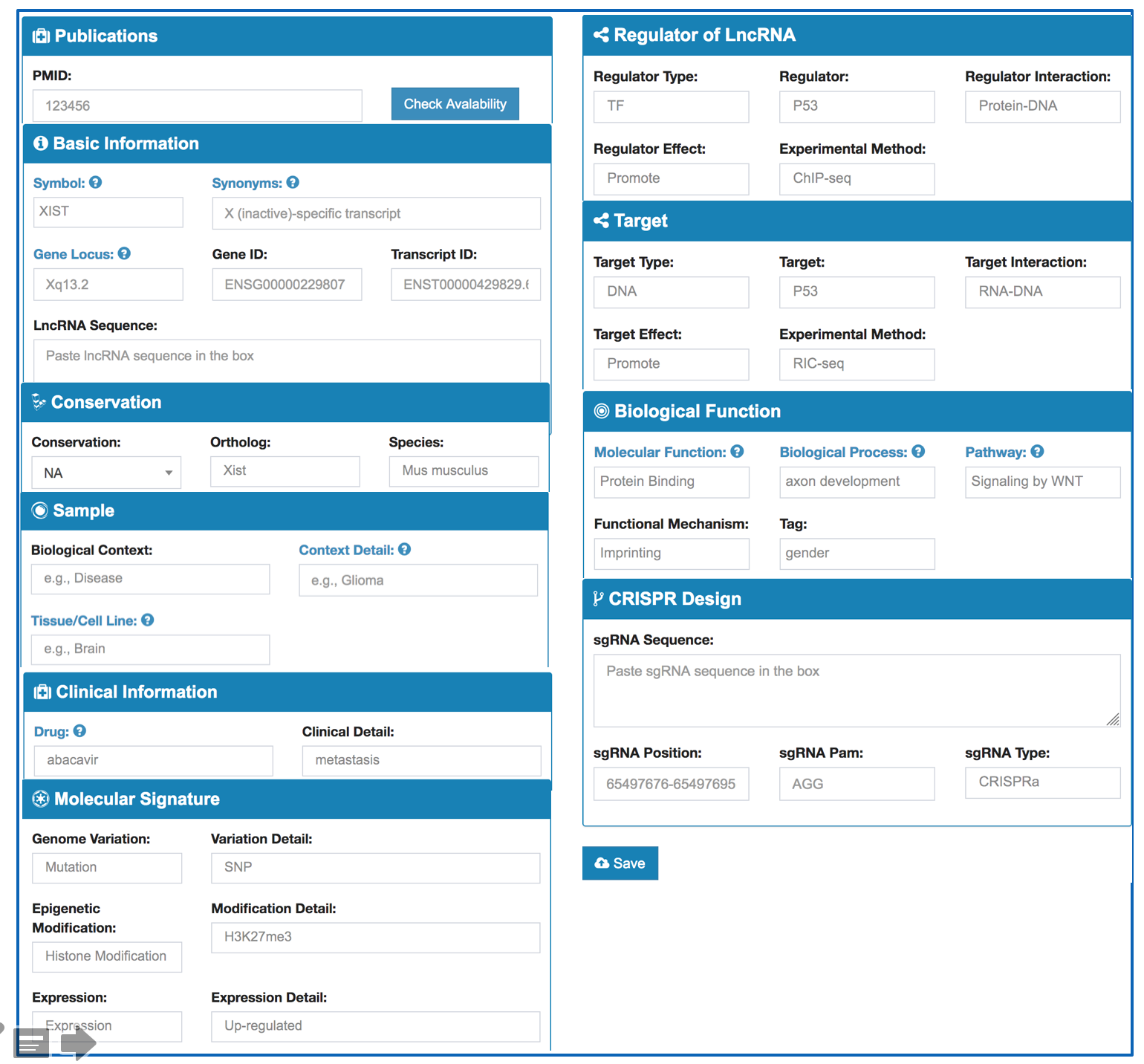
The curation model we used is detailed below.
| Data Type | Description | Value | |
| Basic Information | Symbol | Gene identifier | eg: MALAT1 |
| Synonyms | Gene identifier | eg: LINC00047 | |
| Gene Characteristic | Location of the lncRNA | Gene chromosome | |
| Transcript ID | Transcript identifier | eg: HSALNT0176789 | |
| Transcript Type | The Transcript type | eg: antisense | |
| Conservation | Conservation | True or Yes | eg: Ture |
| Ortholog | Species and conserved lncRNA | Ceg: Human;7SL | |
| Sample | Biological Context | Controlled vocabulary | Aging;Disease;Subcellular Location;Trait;Preimplantation Embryo;Virus Infection;Cell Differentiation;Reproduction;Circadian;Organ Development |
| Context Detail | The name of diseases | eg: breast cancer | |
| Cellular Component | Controlled vocabulary | eg: nucleic | |
| Tissue/cell line | Materials involved in the experiment | eg: MCF-7 | |
| Molecular Signature | Genome Variation | Controlled vocabulary | Locus;Mutation |
| Variation Detail | Detailed variation information of lncRNA | eg:SNP(rs217727) | |
| Epigenetic Modification | Controlled vocabulary | Histone modification;DNA methylation | |
| Modification Detail | Detailed variation information of lncRNA | eg:cg22568540 | |
| Expression | Controlled vocabulary | RNA;Protein | |
| Expression detail | Controlled vocabulary | Down-regulated;Up-regulated;Differentially expressed | |
| Regulator of LncRNA | Regulator Type | Controlled vocabulary | PCG;TF;Protein |
| Regulator | Regulation of lncRNA molecules | eg: MYC | |
| Regulator Interaction | Controlled vocabulary | Protein-DNA;RNA-DNA;RNA-RNA | |
| Regulator Interaction Effect | Controlled vocabulary | promote;inhibit | |
| Target of LncRNA | Target Type | Controlled vocabulary | PCG;TF;protein;mRNA;miRNA;lncRNA;snoRNA |
| Target | Target of lncRNA molecules | eg: mir155HG | |
| Target Interaction | Controlled vocabulary | RNA-RNA;RNA-Protein;RNA-DNA;RNA-TF | |
| Target Interaction Effect | Controlled vocabulary | promote;inhibit | |
| Biological Function of LncRNA | Molecular Function | Controlled vocabulary | eg: GTPase activity |
| Biological Process | Controlled vocabulary | eg:Epithelial-mesenchymal transition | |
| Pathway | Controlled vocabulary | eg:Wnt signaling pathway | |
| Functional Mechanism | Controlled vocabulary | eg:Transcriptional regulation | |
| Tag | Controlled vocabulary | biomarker;epigenetic therapy;hallmark;therapy target;immunity;oncogene;cellular stress;genetic marker;stress action;suppressor;susceptibility gene;transcriptional repressor;molecular scaffold;mediator;hotspot in gwas;prognostic marker;resist ancegene;riboregulator | |
| Clinical Information | Clinical | Controlled vocabulary | metastasis;recurrence;prognosis;drug;survival;clinical;diagnosis |
| Clinical Detail | Clinical information of the lncRNA | eg: TMZ | |
| Experimental Sample | Experiment Method | Method of experimental validation for each lncRNA-target regulation and lncRNA expression | eg: Q-pcr |
| Publication | Reference | Publication in which the interaction is described | PubMed ID |
| Description | Detailed descriptions of the lncRNA in human different context according to the reference | eg: Long noncoding rna afap1-as1 predicts a poor prognosis and regulates non-small cell lung cancer cell proliferation by epigenetically repressing p21 expression. |
1.5 How did we curate data for LncRNAWiki 2.0?
Based on the curation model, we curated lncRNA associations from published literatures, re-organized the annotations in LncRNAWiki 1.0, and collected the annotations from the selected lncRNA databases.
We acknowledge these databases, from which annotations are integrated, RNALocate V2.0, NPInter V4.0, LncRNADisease V2.0, Lnc2Cancer3.0, LncSLdb, Dynamic-BM, LngReg, D-lnc, lncRNA2Target, LncCeRBase, LncR2metasta, EWAS Atlas, LncACTdb 2.0, LncTarD, and CRISPRlnc as well as LncRNAWiki 1.0 .
It is extremely laborious and time-consuming for data integration of different resources, and we hope our curation model will greatly benefit the researchers by easing the efforts for lncRNA curation. Also, we call normalization and standardization for the curation models among different resources.

1.6 How is LncRNAWiki 2.0 organized?
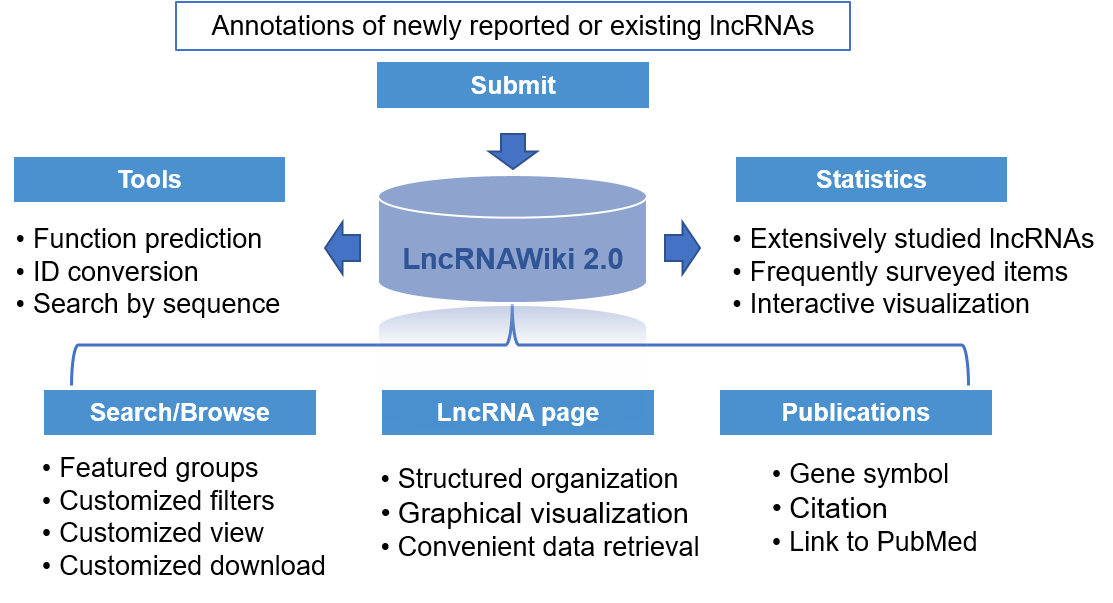
LncRNAWiki consists of six central parts, which are ‘Submit’, ‘Search/Browse’, ‘LncRNA page’, ‘Publications’, ‘Tools’ and ‘Statistics’.
Specifically, ‘Submit’ is provided for data curation, in which registered users could submit data and annotations for any lncRNA(s) of interest. As a result, all lncRNAs and their associations are summarized and presented as a tabular format, which could be browsed by preset groups or with customized filters and easily exported in csv format in ‘Browse’. Additionally, detailed annotations for each lncRNA are presented in a structured manner and publicly accessible and downloadable in ‘LncRNA page’. Also, related publications of each lncRNA are archived in ‘Publications’.
Based on the standardized curation model, abundant controlled vocabularies, and comprehensive data, LncRNAWiki presents a series of data statistics, which would keep the researchers informed of the progresses and hotspots of lncRNA studies. Moreover, to facilitate users to identify lncRNAs with potentially important functions, function prediction is achieved by associating with curated interacting partners (regulators and targets), as well as co-expressed genes sourced from LncExpDB.
1.7 How does LncRNAWiki 2.0 ensure the high-quality curation?
To ensure high-quality curation, expert curators are recruited to review and check these submissions/edits, and only reviewed annotations with literature support can be incorporated into LncRNAWiki 2.0.

Notably, any user could report errors in the lncRNA page, which can be achieved conveniently just by clicking on ‘Report’ in each section without registration. When any error report is sent out, LncRNAWiki 2.0 is able to automatically notify expert curators to review and check the reported error, with the aim to ensure curation quality in LncRNAWiki 2.0.

2. Curation and Account
2.1 How to acquire a LncRNAWiki 2.0 account?
By clicking on ‘Sign in’ on the top right of the homepage, any registered user will be asked to enter the email and password to login. If not registered, users should register first with their emails, personal information, and institutional information.
2.2 Is it allowed for any user to provide edits in LncRNAWiki 2.0?
LncRNAWiki 2.0 allows any user to view and search but only registered users can add and edit content.
2.3 Why can’t I log into LncRNAWiki 2.0?
* Make sure that the Caps Lock key is not depressed. Passwords are case sensitive.
* Make sure your browser is set to accept cookies.
* Contact us at lncwiki@big.ac.cn.
2.4 How to submit a lncRNA?
To ensure quality of community-curated annotation items, we have provided a curation handbook to introduce the standards and examples for each subject, which could be downloaded here .
Click on ‘Submit’ after login, and then input the lncRNA information in the submission page based on the structured curation model. Based on a publication, input basic information, conservation, sample, clinical information, molecular signature, regulator of lncRNA, target, biological function of lncRNA, publications and then click on ‘submit’. For any individual submission, please note that the submitted annotations should be derived from one single publication.

2.5 How can I edit or update the annotation of a lncRNA?
After login, click ‘My account’ to access the curator page and to view all the curation records. Choose the lncRNA you have submitted and click ‘Edit’ to edit or update the related information.

3. Usage
3.1 How to download the data?
In the Browse page, users can download the results with customized filters.

In the lncRNA page, users can download the data of interest by clicking ‘CSV’ or ‘Copy’ nearby each table.

3.2 How to get the function of interested lncRNAs?
- Function associations curated from literatures are available in each lncRNA page, in ‘Manual Curation’ of the Biological Function section.
- Function predictions are performed based on interacting partners, and tables and figures could be accessed in ‘In Silico Prediction’ of the Biological Function section, including GO terms of molecular function (MF), biological process (BP), cellular component (CC) and KEGG pathway. In addition, in silico predictions could be accessed through ‘Functional Prediction (Curated interacting partners)/Function Prediction (Co-expression mRNAs)’ in ‘Tools’.
3.3 What kind of tools can I use in LncRNAWiki 2.0?
LncRNAWiki 2.0 provides three tools, which are developed for Functional Annotation, ID Conversion and BLAST, respectively.- Functional Annotation: Users can input the lncRNA symbol and get the associated GO terms including molecular function (MF), biological process (BP), cellular component (CC) and KEGG pathways. Biological function is predicted based on the interacting partners derived from manual curation and co-expressed genes sourced from LncExpDB.
-
ID Conversion: LncRNAWiki 2.0 provides the ID conversion between gene symbol and accession ID across different databases including LncExpDB, GENCODE, LNCipedia, NONCODE, BIGtranscriptome, CHESS, RefLnc, MiTranscriptome, and FANTOM. Users can convert the gene symbol into gene/transcript IDs.
- BLAST: Users can search lncRNAs by sequences using BLAST.

We perform ID conversion based on the lncRNA integration results of LncExpDB [PMID: 33045751]. Related sequences across different databases are defined as those with exact matches of all exon junctions as well as 5'-start and 3'-end boundaries. Specifically, the similar transcripts were identified with the comparison code ‘ = ’ of Gffcompare (means complete match of intron chain), and 5'-start/3'-end boundaries were further compared with in-house scripts. In addition, questionable lncRNAs including the potential protein-coding RNAs, incomplete and unreliable transcripts, background noises in each database were removed.


3.4 How to cite LncRNAWiki 2.0?
LncRNAWiki 2.0: a knowledgebase of human long non-coding RNAs with enhanced curation model and database system. (In preparation)
3.5 Licenses?
LncRNAWiki 2.0 is free for academic use only. For any commercial use, please contact us for commercial licensing terms.