Tutorial
1. Genes
Genes provides the gene information of the sheep genomes. For searching gene data, filter conditions are circled by yellow boxes as Figure 1-1 showed:
- Gene position
- Gene type
- Gene name
- Gene symbol
Detail information and variant information in this gene could be obtained if users click gene name, like 100034665. While users click gene symbol, the website of Ensembl would be opened and the result of this gene would be showed.

Figure 1
2. Variants
A total of 82,689,498 genomic variants (including 70,370,968 SNPs and 12,318,530 small INDELs) were called from 355 sheep of whole genome sequence, merging into 1512 sheep of Illumina Ovine SNP50K BeadChip(50K) and 911 sheep of llumina Ovine Infinium HD SNP BeadChip(600K). For searching variants data, filter conditions are circled by yellow boxes as Figure 2-1 showed:
- Name: Variant ID, Gene name, dbSNP
- Position
- Variant type: SNP, INDEL or ALL
- Technology: WGS, 600K and 50K
- Consequence type (the annotation results of VEP)
- MAF

Figure 2-1
Detail information including basic information, gene annotation, genome browse and the genotype of 355 WGS individuals would be obtained while users click the specific variant ID (eg: oas1883). Meanwhile, the genotype of this variant in SNP chip can also be gotten if exists.

Figure 2-2
3. Breeds
"Breeds" provides breed information of sheep. It was collected from public resources. For searching target breed, filtering conditions are circled in yellow as showed in Figure 3:
- Breed name
- Breed country
- Use
- Bodysize
- Weight males/femles
Detailed information will be shown while clicking the specific breed name.
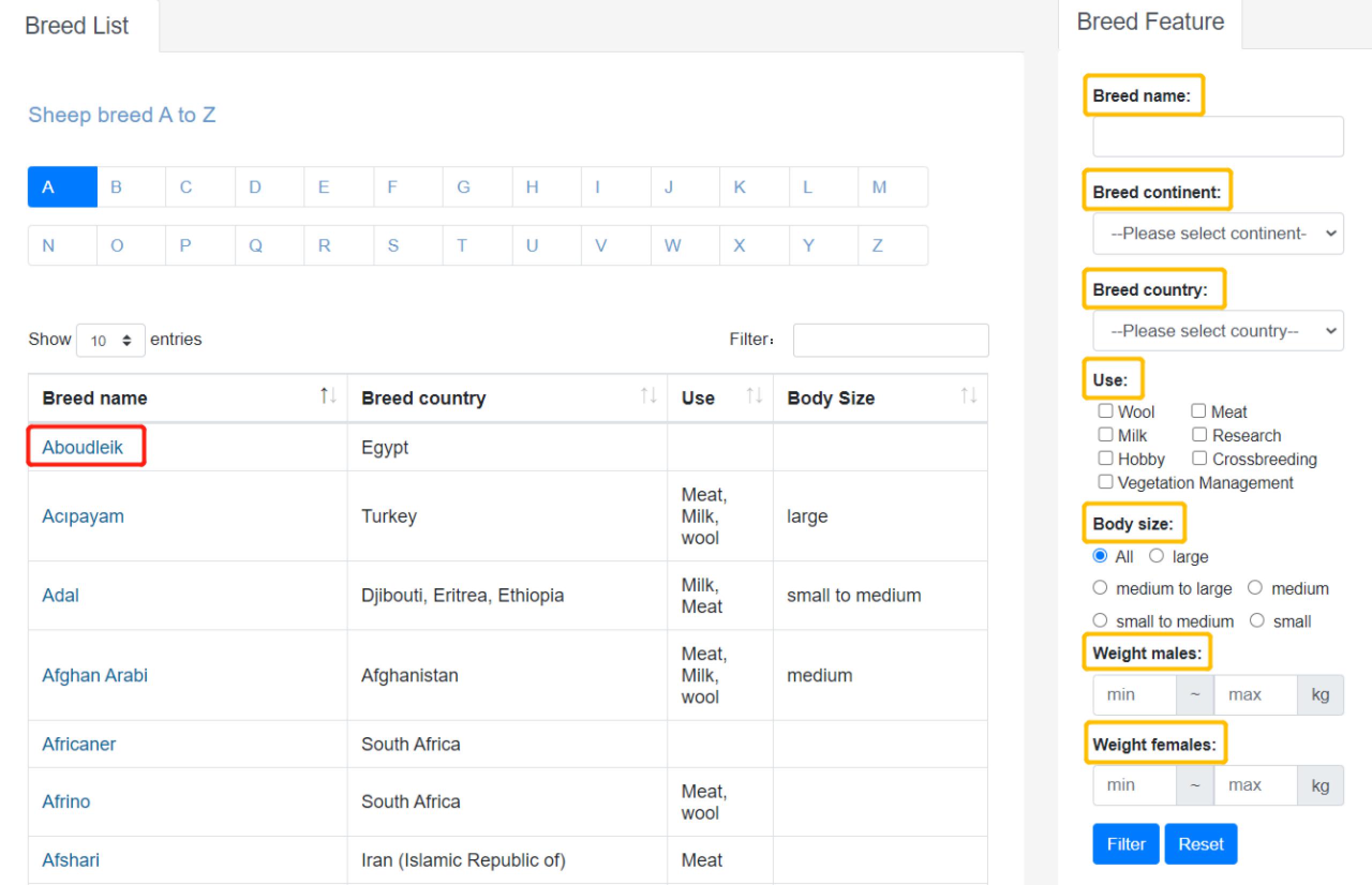
Figure 3
4. Associations
"Associations" contains 911 variants excavated from 52 papers published bewteen 2011 and 2019. For searching genotype or phenotype, filtering conditions are circled in yellow as showed in Figure 4.
- Variant ID
- dbSNP ID
- Position
- Trait
The front-page of variant information in iSheep, EMBL-EBI (the European Bioinformatics Institute), or the PubMed page of associated paper will be opened by clicking Variant ID, dbSNP ID, or PubMed ID. A related .csv file containing filtered results will be obtained when clicking the “CSV” button.

Figure 4
5. Publications
"Publications" includes 52 papers which used for G2P(genetype to phenotype) associations above. And they were divided into five parts due to their trait type (Figure 5). Click the num on the right of the different trait type and the results would be showed.
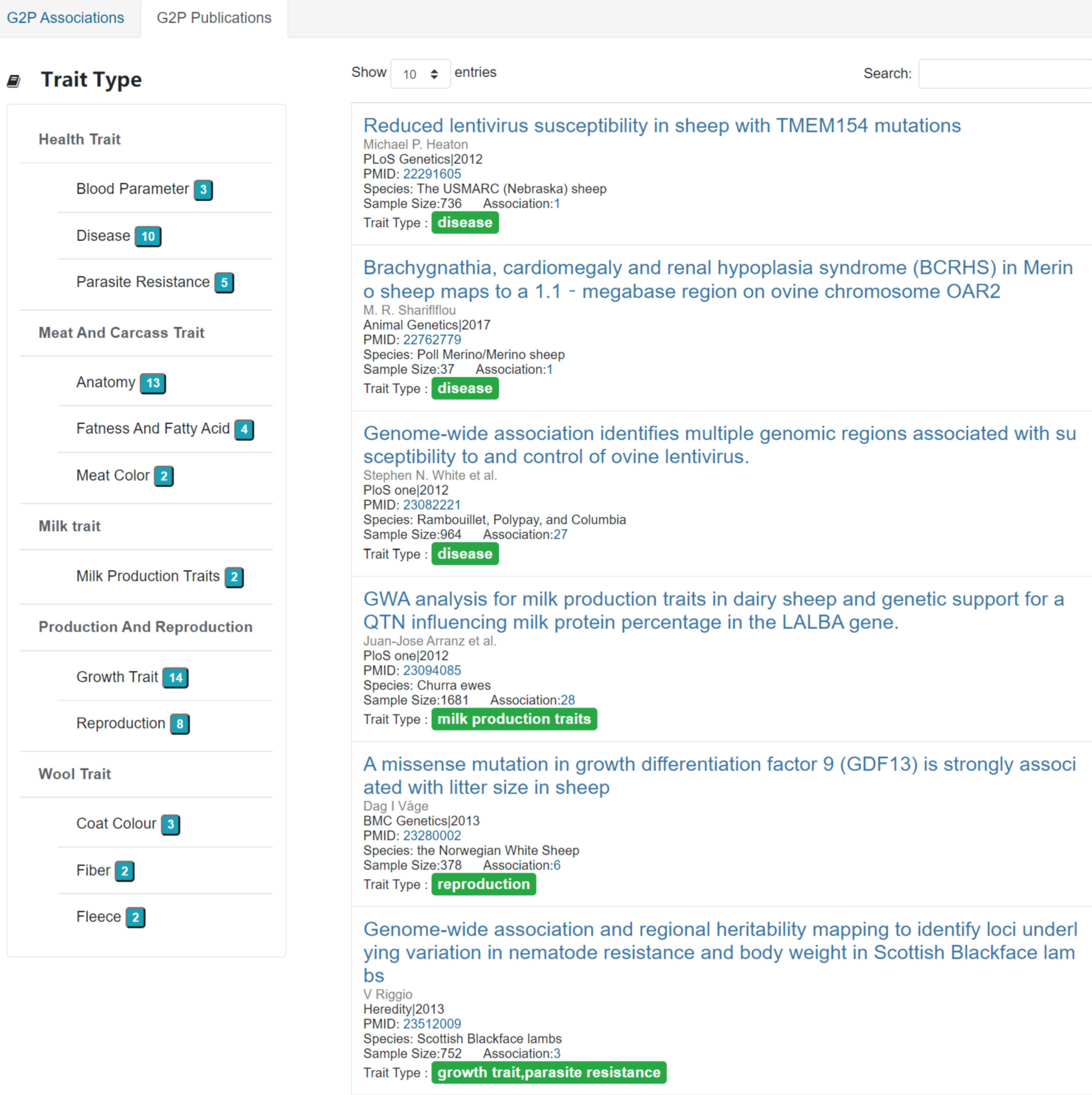
Figure 5
6. Samples
"Samples" provides the sample information of WGS, Illumina Ovine SNP50K BeadChip and Illumina Ovine Infinium HD SNP BeadChip data. Click the Biosample ID, Bioproject ID or Breed name, the related detail information will be showed, separately. Raw WGS data have been deposited in NCBI SRA and NGDC GSA. Users can jump to associated pages via this part if they want to download them.

Figure 6
7. Comparison
"Comparison" supports users to search common SNPs between two or more individuals. After selecting individuals, position, consequence type and associated genes could be used for filtering(Figure 7).
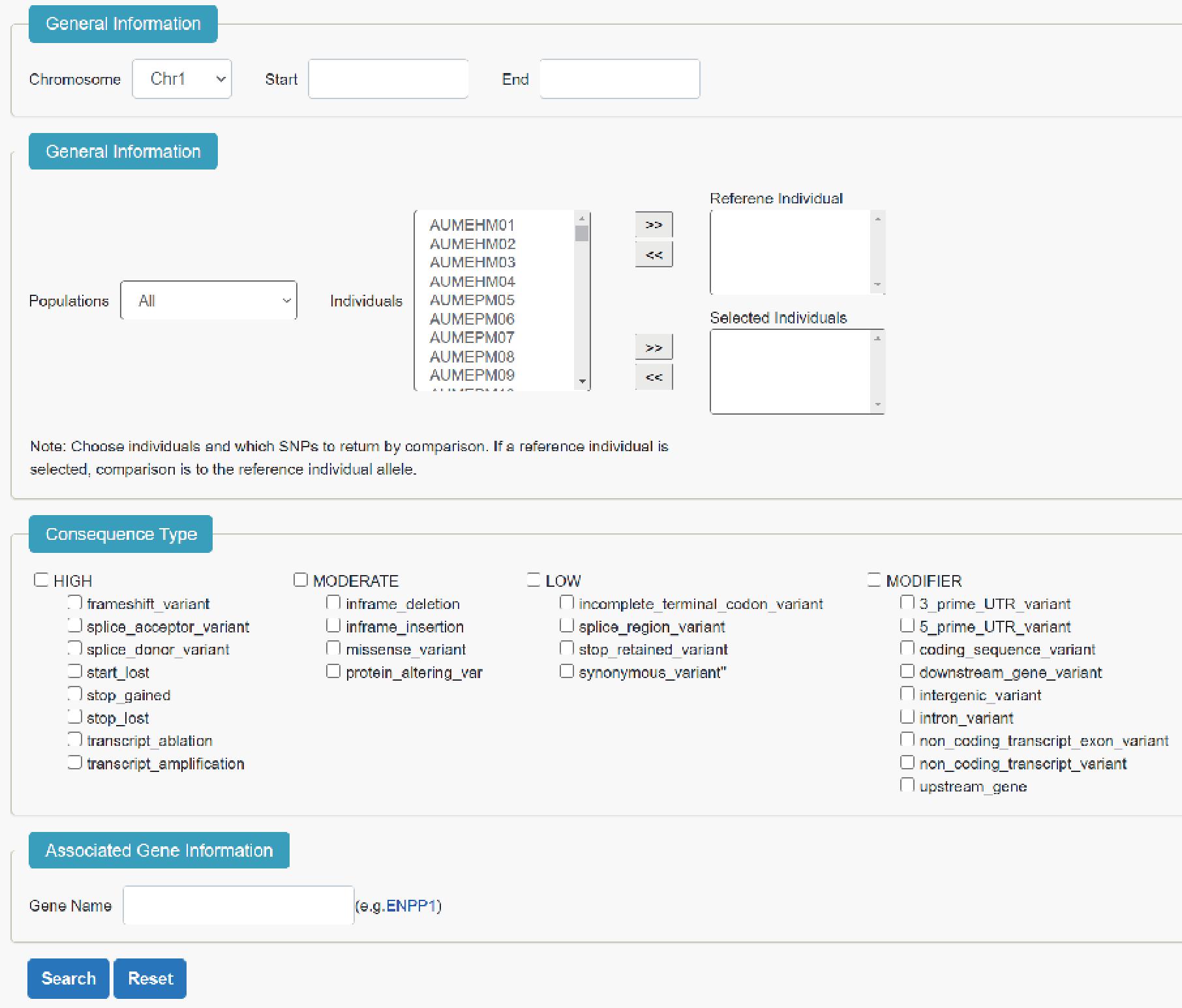
Figure 7
8. Download
"Download" consists of two parts: (a) iSheep data includes Illumina Ovine SNP50K BeadChip and Illumina Ovine Infinium HD SNP BeadChip variant calling file, 355 WGS INDELs and SNPs data; (b) reference genome and annotation data includes ten reference genomes about five Ovis species.
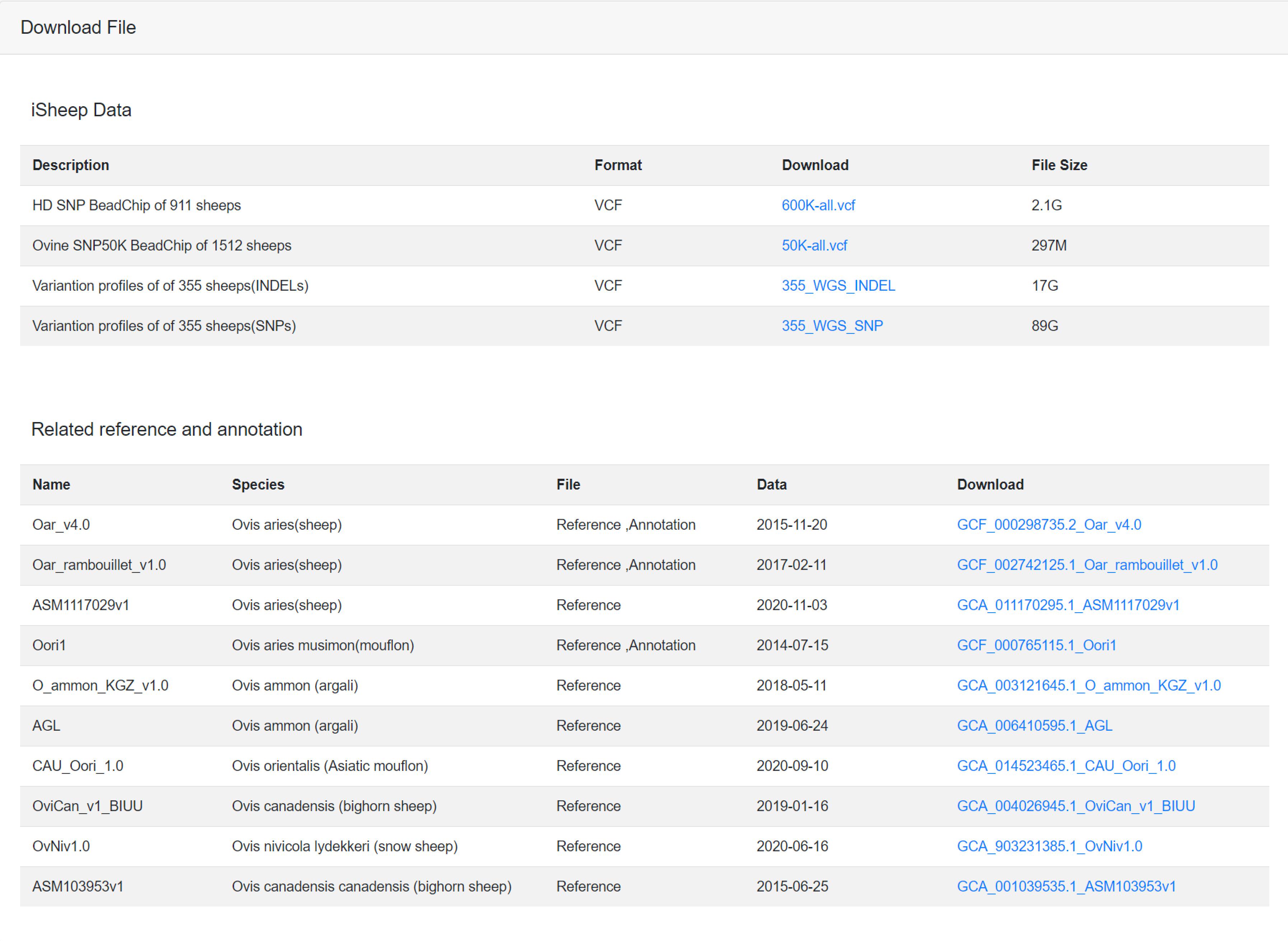
Figure 8