FAQ
The Genome Warehouse website and database provide you with access to genome data for 27 released plant and animal genomes. Sequence similarity (BLAST) and keyword-based searching can help you pinpoint genes of interest in corresponding species. Genome browsing is available via our deployment of JBrowse. Our JBrowsers include gene tracks. Finally, all data on genes and individual genomes in the Genome Warehouse database are downloadable via FTP.
The Home Page
Genome Warehouse's home page provides a quick search interface for learning about the genomes included in the current release and finding genes on interest. Release Notes and News on the home page for the current release provide information on: what's New in the current release, in terms of features, data, and bug fixes; which genomes in which versions are included. This is another quick way to find out about new genome updates. There's a single search interface for keyword-based searches, accessible by selecting "keyword search" on the home page. The search interface lets you efficiently select species targets by typing the names of your desired search targets. Enter one or more search times that match gene identifiers, controlled ontology terms, gene symbols, etc.
Finding Out Genomes in Genome Warehouse
Genomes are organized into three groups: plant genomes, animal genomes and all released genomes. You can switch
between groups by clicking on the ‘Species’ on the top menu button, the pull-down menu ‘All Species’ or the ‘Tree
View’ will lead to individual species on interest. The list of genomes is hyperlinked to individual organism
pages, each genome has an information organism page. Here is the information for Vitis vinifera (Figure 1).
 Figure 1. Basic genomic information page for Vitis vinifera
Figure 1. Basic genomic information page for Vitis vinifera
Genomes will always list a "Data Source", linked out to the institutional or group website from which the data
was obtained. Next comes a brief overview of the genome, and summary of assembly and annotation statistics, a
section on "Sequencing, Assembly and Annotation", a project contact list, and a reference publication (for
published genomes) that should always be cited when using the data from this project (Figure 2).
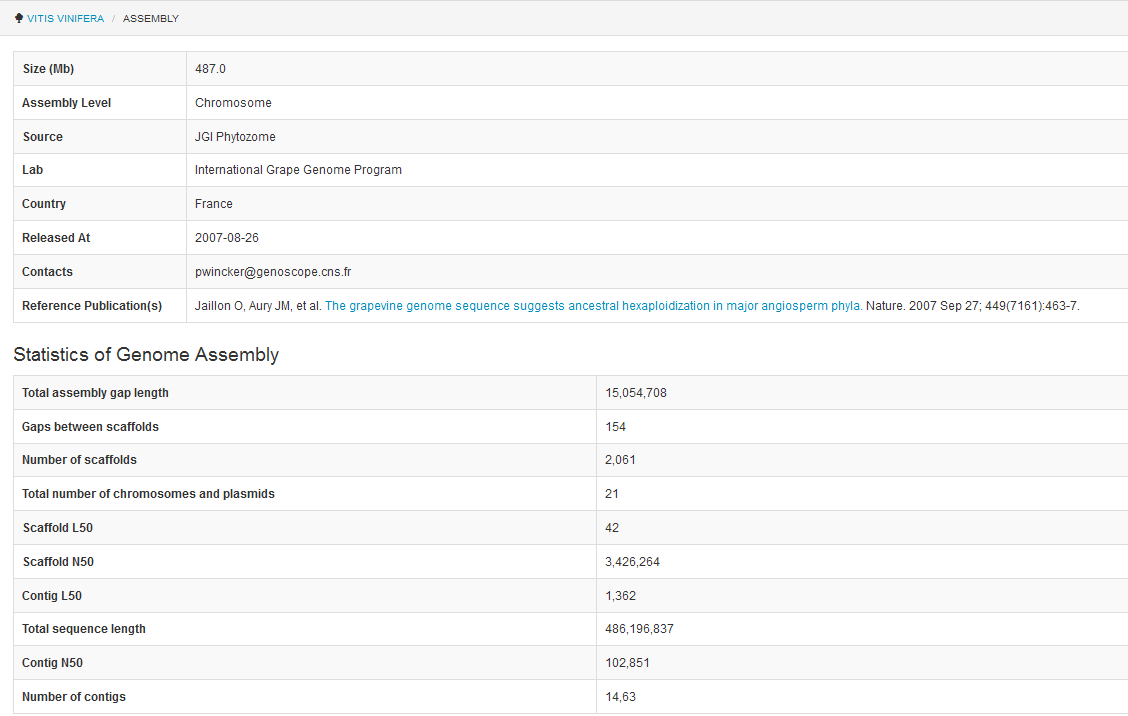 Figure 2. Summary of assembly and annotation statistics
Figure 2. Summary of assembly and annotation statistics
The BLAST Search Interface
The "BLAST Search" buttons will take you to the main search interface page, with that search tool selected, and the search target set to the corresponding organism. The method to build your query:
- The "BLAST Search" buttons will take you to the main search interface page, with that search tool selected, and the search target set to the corresponding organism. The method to build your query:
- Decide whether you want to view BLAST results in the browser, or prefer email notification of job completion (Note: you MUST select this option for uploaded multifasta query sequences, as browser rendering of multi-sequence query jobs is not supported).
- Enter your email address if necessary (if notification was chosen above).
- Make sure you choose the target type (Genome or Proteome) using the Target Type pulldown.
- Choose the BLAST program based on your goal, and the particular combination of query and target sequence types.
- Tune other NCBI blast parameters as desired, or leave at their default values.
- Click the green "GO" button at top and wait for results.
Viewing Genomic Data in JBrowse
The JBrowse link will take you to a default genomic view in the JBrowse genome browser for this organism, from where you can browse tracks or search for features in that context.
Download Bulk Data Files
The "Download" button will take you to the bulk data files for all genomes deposited in the Genome Warehouse. Access to bulk data files for each species is made available via the BIGD FTP.
